Research
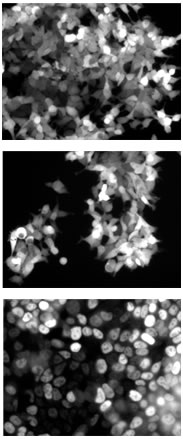
| Research Synthetic and quantitative biology, DNA nanotechnology, RNA-based mechanisms for gene regulation, engineering of molecular circuits. Biological organisms process information and control cellular behavior using sophisticated biochemical circuits. In order to engineer circuits of similar complexity and thus “program” biology we need to develop molecular tools for (i) accurate detection of complex cellular states and (ii) control and modulation of those states. Our lab takes a comprehensive approach for solving these problems using nucleic acid-based molecular circuitry. Nucleic acids play a dual role in biology: their sequences and expression levels store in- formation about the state of a cell and regulatory RNAs also actively influence that state. MicroRNAs (miRNAs) are now believed to regulate around 30% of all protein-coding genes in humans. Nucleic acids are an ideal substrate for rationally designing synthetic regulatory elements, because their interactions can be predicted from their sequence. In vitro nucleic acid circuits. We have implemented multi-layered nucleic acid logic circuits that function reliably in an aqueous, cell-free environment. Hybridization reactions provide the free energy to move computation forward and Watson-Crick base pairing between modular recognition domains determines the connectivity of circuit components. Circuits embody key design principles of digital electronics: logic, cascading, restoration, fan-in/fan-out and modularity. Our approach demonstrates that adherence to these principles provides a viable path towards the de novo construction of biochemical reaction networks. Estimates of the size of a minimal genome range from about 50 to around 450 genes. Thus, synthetic biochemical reaction networks with just a few hundred components would approach the computational complexity (although not necessarily the chemical variety) of the circuitry required for a simple biological organism. It is likely that such complexity can be reached using only nucleic acid components. Working in vitro it is possible to rapidly scale up circuit complexity, to explore new modes of molecular computation, and to quantitatively characterize circuit behavior without degradation of DNA or RNA that may occur in a cell. These experiments represent a foundation for developing in vivo computation using related molecular schemes. RNA tools for conditonal gene regulation In order to program biological systems we need to develop molecular components that work reliably in a cellular environment. We are currently working on giving proof of principle that synthetic nucleic acid logic gates can detect mRNA and microRNA inside cells and, in response to varying expression patterns, can differentially regulate target genes.Target regulation is achieved by coupling synthetic circuitry to existing cellular pathways like the RNAi pathway. Synthetic nucleic acid systems for conditional gene regulation have potentially interesting application as "smart drugs" since they allow to restrict gene knockdown to a subset of cells characterized by a specific gene expression pattern. We are characterizing circuit properties quantitatively and on the single cell level using fluorescence time-lapse microscopy techniques and cultured mammalian cells. Quantitative understanding is required for optimizing component designs and for subsequent construction of multi-component circuits. Re-engineering the RNAi/microRNA pathway. The miRNA pathway provides an ideal starting point for genetically engineering synthetic regulatory networks that are fully integrated with the cellular environment. MicroRNA-based gene regulation relies on Crick-Watson base pairing and is relatively predictable. We also know how to add new miRNAs to a cellular network or how to inactivate existing ones. We want to demonstrate that synthetic miRNA circuits are capable of guiding complex cellular processes like differentiation. Such an engineering approach will further our understanding of eukaryotic gene regulation. As a first step in this direction we are currently also investigating the dynamics of biological microRNA. ©2009 University of Washington. All Rights Reserved. last update: 03/13/2009 |