

Genome-wide MyoD Binding in Skeletal Muscle Cells: A Potential for Broad Cellular Reprogramming
Yi Cao,1,7 Zizhen Yao,2,7 Deepayan Sarkar,2 Michael Lawrence,2 Gilson J. Sanchez,1,4 Maura H. Parker,3 Kyle L. MacQuarrie,1,4 Jerry Davison,2 Martin T. Morgan,2 Walter L. Ruzzo,2,5 Robert C. Gentleman2,* and Stephen J. Tapscott1,3,6,*
Developmental Cell. 2010 Apr 20;18(4):662-74. doi:10.1016/j.devcel.2010.02.014 pmid: 20412780
Comment: Biggin MD. "MyoD, a lesson in widespread DNA binding." Dev Cell. 2010 Apr 20;18(4):505-6. doi:10.1016/j.devcel.2010.04.004 pmid: 20412764
 SUMMARY
Recent studies have demonstrated that MyoD initiates a
feed-forward regulation of skeletal muscle gene expression, predicting
that MyoD binds directly to many genes expressed during
differentiation. We have used chromatin immunoprecipitation and
high-throughput sequencing to identify genome-wide binding of MyoD in
several skeletal muscle cell types. As anticipated, MyoD
preferentially binds to a VCASCTG sequence that resembles the in
vitro-selected site for a MyoD:E-protein heterodimer, and MyoD binding
increases during differentiation at many of the regulatory regions of
genes expressed in skeletal muscle. Unanticipated findings were that
MyoD was constitutively bound to thousands of additional sites in both
myoblasts and myotubes, and that the genome-wide binding of MyoD was
associated with regional histone acetylation. Therefore, in addition
to regulating muscle gene expression, MyoD binds genome wide and has
the ability to broadly alter the epigenome in myoblasts and myotubes.
SUMMARY
Recent studies have demonstrated that MyoD initiates a
feed-forward regulation of skeletal muscle gene expression, predicting
that MyoD binds directly to many genes expressed during
differentiation. We have used chromatin immunoprecipitation and
high-throughput sequencing to identify genome-wide binding of MyoD in
several skeletal muscle cell types. As anticipated, MyoD
preferentially binds to a VCASCTG sequence that resembles the in
vitro-selected site for a MyoD:E-protein heterodimer, and MyoD binding
increases during differentiation at many of the regulatory regions of
genes expressed in skeletal muscle. Unanticipated findings were that
MyoD was constitutively bound to thousands of additional sites in both
myoblasts and myotubes, and that the genome-wide binding of MyoD was
associated with regional histone acetylation. Therefore, in addition
to regulating muscle gene expression, MyoD binds genome wide and has
the ability to broadly alter the epigenome in myoblasts and myotubes.
DATA AVAILABILITY Raw data have been deposited in the Sequence Read Archive (SRA), accessions SRP001761 and SRA010854, and in the Gene Expression Omnibus, accession GSE20059.
As a supplement to the raw SRA data, this web site provides processed results in formats suitable for viewing as "Custom Tracks" in the UCSC Genome Browser. Each data set is a browser "wiggle" track, provided in two different formats: a (gzip-compressed) .bedGraph file, and a .bigWig file. Download these files if desired (use the "Download" links), but most are very large. Conveniently, the .bigWig format allows the files to be viewed in the browser without downloading them to your computer. (Use the "View in Browser" links. Return and click additional ones to see several at once; the browser retains your preferences.) The .bedGraph files contain the same data. Most are too large to upload to UCSC, but this format may be more convenient for other analyses.
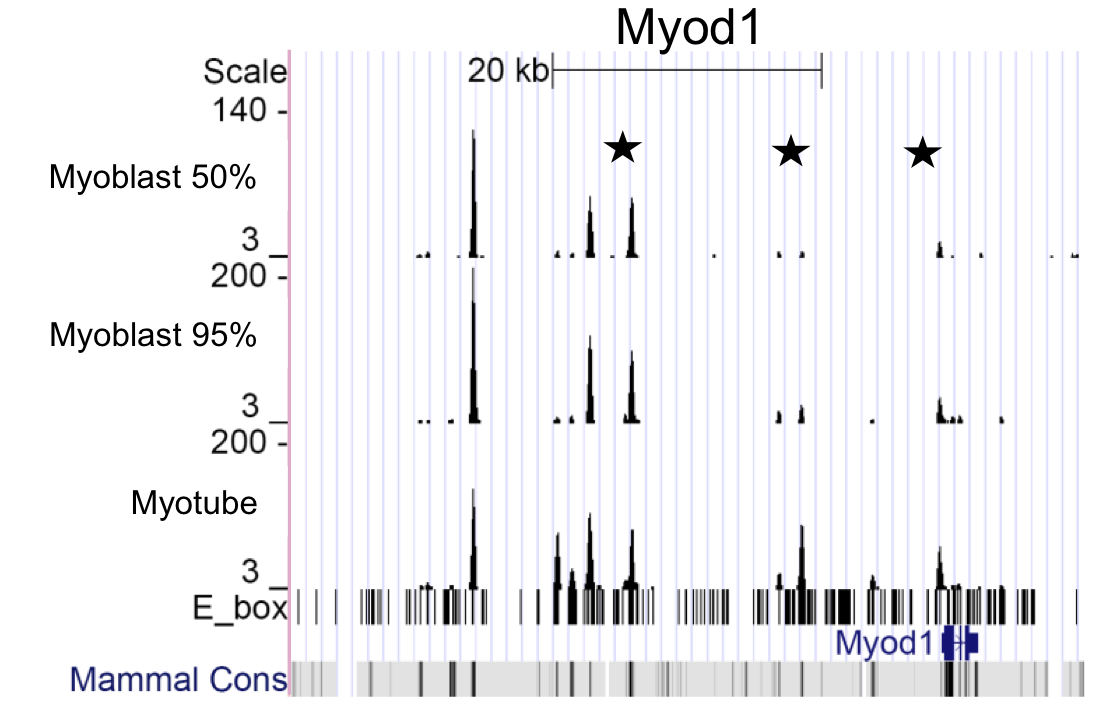
The Summary datasets below give the coordinates and heights of the 23,271 and 25,956 peaks of height 12 or greater found in 95% confluent C2C12 myoblasts and myotubes, resp. (A "peak" is a maximal contiguous region of coverage at least 4 surrounding a region of coverage at least 12; the height of that peak is the maximum coverage found in that interval.)
The Full datasets give the coverage (locations and cummulative heights) of all Illumina Genome Analyzer II reads in our MyoD ChIPseq experiments mapped to the mouse mm9 genome assembly. The five experiments reported here are C2C12 myoblasts (50% and 95% confluency), C2C12 myotubes, mouse embryonic fibroblasts transfected with MyoD via a retroviral vector, and primary mouse myotubes.
In all cases, "coverage" is defined by extending each mapped read in the sequencing orientation to a total of 200bp in length and counting the number of extended reads that cover each position in the genome. Reads from both genomic strands are combined.
| Summary Datasets | ||||||
| C2C12 Myoblasts, height >= 12 | View in Browser | Download: | blast .bigWig (336K) | .bedGraph.gz (218K) | ||
| C2C12 Myotubes, height >= 12 | View in Browser | Download: | tube .bigWig (507K) | .bedGraph.gz (242K) | ||
| Full Datasets | ||||||
| C2C12 Myoblasts, 50% confluence | View in Browser | Download: | myoblast50 .bigWig (147M) | .bedGraph.gz ( 39M) | ||
| C2C12 Myoblasts, 95% confluence | View in Browser | Download: | myoblast95 .bigWig (209M) | .bedGraph.gz ( 60M) | ||
| C2C12 Myotubes | View in Browser | Download: | myotube .bigWig (185M) | .bedGraph.gz ( 51M) | ||
| Mouse embryonic fibroblasts + MyoD | View in Browser | Download: | fibroblast .bigWig (725M) | .bedGraph.gz (195M) | ||
| Mouse primary myotubes | View in Browser | Download: | primary.myotube .bigWig (324M) | .bedGraph.gz ( 78M) | ||
If you have problems with this web page, please contact ruzzo![]() uw.edu.
uw.edu.
1Human Biology Division
2Public Health Sciences Division
3Clinical Research Division Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA
4Molecular and Cellular Biology Graduate Program
5Departments of Computer Science and Engineering and Genome Sciences
6Department of Neurology University of Washington, Seattle, WA 98105, USA
7These authors contributed equally to this work
*Correspondence: rgentlem![]() fhcrc.org (R.C.G.), stapscot
fhcrc.org (R.C.G.), stapscot![]() fhcrc.org (S.J.T.)
fhcrc.org (S.J.T.)