Research
The rapidly growing availability of high-throughput biological data (such as microarray, RNA-seq, exome sequence, epigenomic profiles) has opened new opportunities for addressing key questions in biology and improving health care. Success will increasingly depend on an effective computational methodology that can integrate heterogeneous data sets, effectively reduce the dimensionality of data, extract meaningful patterns out of a number of irrelevant features, and integrate across fields of biology. More and more of biological and medical sciences is becoming an information science.
My group focuses on developing such computational methods based on machine learning and statistics. Specific research projects can be divided into the following categories:
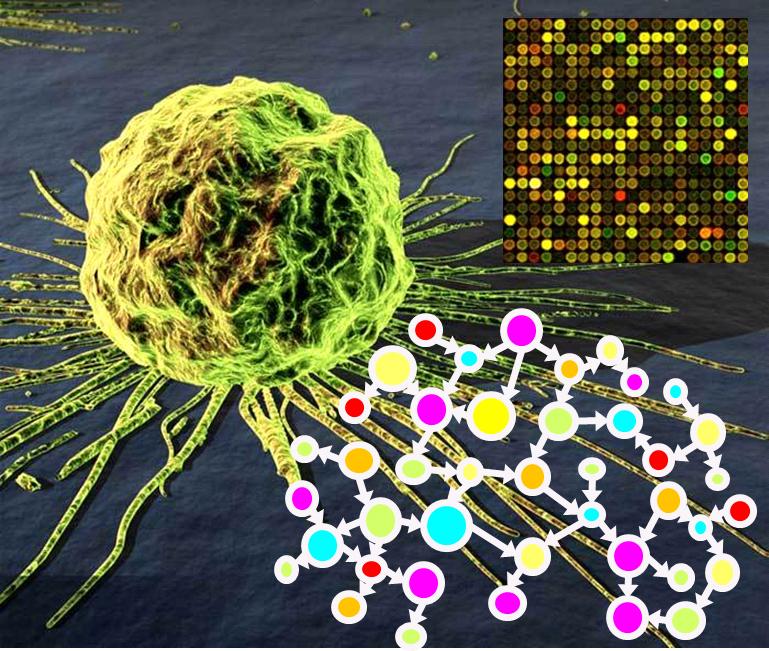
Functional genomics towards personalized health care
 |
more coming soon! |
Collaborators: Anthony Blau (UW Medicine), Pam Becker (UW Medicine), Ray Monnat (UW Pathology), Chris Miller (UW Medicine), Fred Hutchinson Cancer Research Center, and Cardiovascular Health Research Unit
Systems biology of disease mechanisms, complex biological traits, development, evolution and tissue specificity
Collaborators: Stanford Center for Cancer Systems Biology, Daniela Witten (UW Biostatistics), Jake Lusis (UCLA), Tom Drake (UCLA), Daphne Koller (Stanford), Andrew Gentles (Stanford), Maitreya Dunham (UW Genome Sciences)
Systems genetics – system-level understanding of the effect of genetic/epigenetic variation on complex traits
|
|
Collaborators: Aimee Dudley (ISB), Daphne Koller (Stanford), Dana Pe'er (Columbia)
Genomics meets genetics – integrating functional genomic data to inform genome-wide association studies
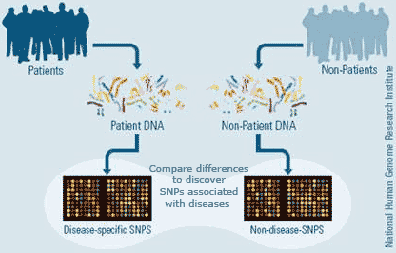 |
Pathway-based genome-wide association studies (GWAS) |
Collaborators: Greg Cooper (HudsonAlpha), Sina Gharib (UW Medicine), Nona Sotoodehnia (UW Medicine)
Predictive medicine
 |
Collaborators: Meliha Yetisgen-Yildiz (UW Biomedical Informatics), Heather L. Evans (UW Medicine)
Developing novel machine learning and data mining techniques
 |
feature learning, transfer learning, sparse structural learning, feature selection |
Collaborators: Daniela Witten (UW Biostatistics), Ruslan Salakhutdinov (U of Toronto), Honglak Lee (U of Michigan)
Collaborations
Our lab has close collaborations with UW Medicine, Genome Sciences, Epidemiology, UW Medical Center, Stanford Center for Cancer Systems Biology, Fred Hutchinson Cancer Research center, Allen Institute for Brain Sciences, and Seattle Children's Hospital. This enables us to access to various types of high-throuput data including genotype, microarray, RNA-seq, exome-sequencing, proteomic data and various epigenomic profiles. The short-distance collaborations also facilitate experimental or clinical validation of the hypotheses generated by our computational models, which will amplify the impact of our approaches.
Systems biology
Learning time-varying regulatory networks for understanding developmental regulation of neuron morphogenesis
In collaboration with Jay Parrish (UW Biology) and Charlie Kim (UCSF)
More coming soon!
Inferring biological processes from gene expression data
In collaboration with Serafim Batzoglou (Stanford)
|
|
Individual genetic variation and gene regulation
Learning the regulatory potential of sequence variations
In collaboration with Aimee Dudley (ISB), David Drubin, Pamela Silver (Harvard), Nevan Krogan (UCSF), Dana Pe'er (Columbia), Daphne Koller (Stanford)
|
|
Reconstructing genetic regulatory networks from eQTL data
In collaboration with Dana Pe'er (Columbia), Aimee Dudley (ISB), George Church (Harvard) , Daphne Koller (Stanford)
|
|
|
Sparse structure learning
Learning a meta-level prior for feature relevance from multiple related tasks
In collaboration with Vassil Chatalbashev, David Vickrey and Daphne Koller (Stanford)
|
|
|
Efficient structure learning of Markov networks using L1-regularization
In collaboration with Varun Ganapathi, Daphne Koller (Stanford)
|
|
|
Efficient learning of L1 Regularized Logistic Regression
In collboration with Honglak Lee, Pieter Abbeel, Andrew Ng (Stanford)
|
|
|